The topic of my Msc dissertation is “Investigating AI and Digital Pathology” which mainly uses the Deep Learning to detect mitosis. This post I will introduce the techniques of the Digital Pathology.
Cancer is one of the main causes of deaths impacting not only human but also animals. This diagnosis of cancer is based on the histological slides of the body tissues. In the traditional pathology, microscope is used as a major tool to review the tissues on the physical glass slides. The analysis of the histological images is usually performed by the pathologists which is an exhaust and tedious task. Digital pathology is a sub-field of the pathology that converts the traditional slides into the digitalized images.
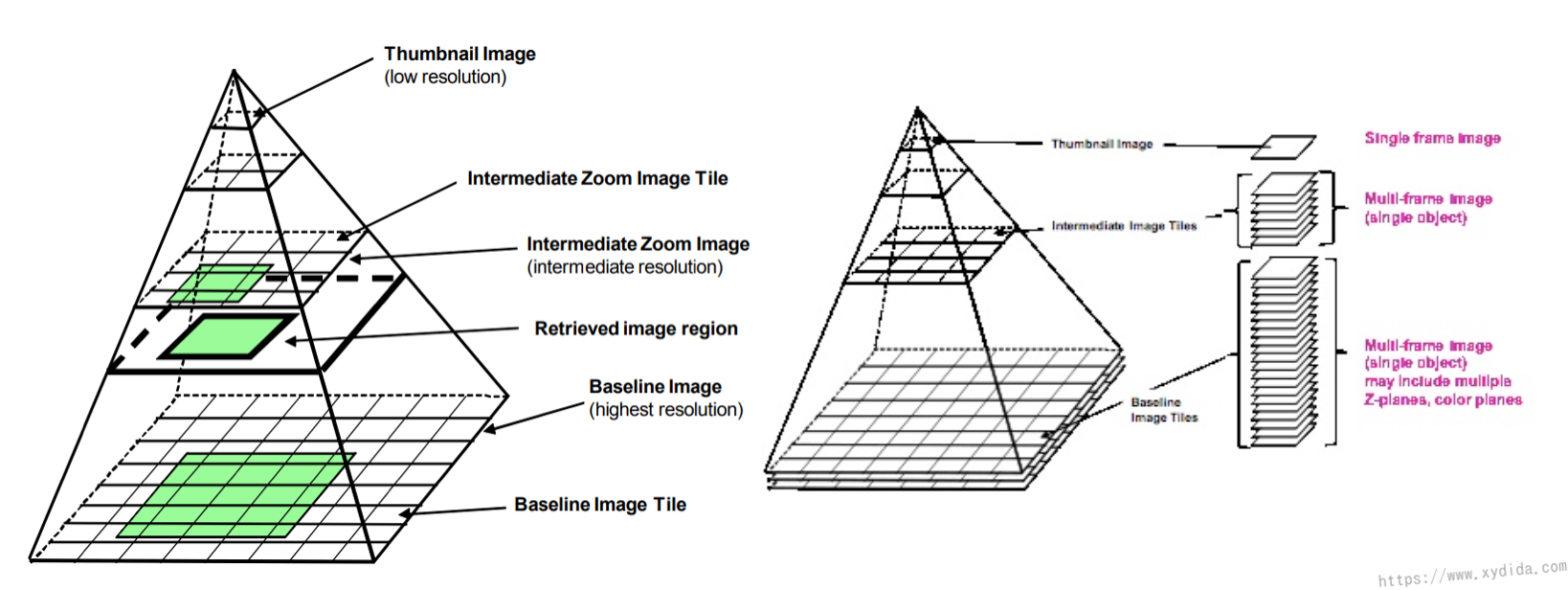
Digitalized images are also called “Whole-Slide Images (WSI)” that are produced by the high-resolution slide scanners at a very high level maginifications such as 20x or 40x. A slide is divided into several stripes and combined as a whole-slide images after scanned. WSI is an uncommon image format such as ndp, svs that can not be viewed or edited by the common softwares. Due to the high quality of the WSI, one digitalized image often has a large data volumn. In my study, the max size of the WSI is 2.3GB, the average size of the WSI is over 1GB as well and the 11 total WSI have a size over 16GB. Thus, the organization of the WSI is different from the common images Fig. 2 shows the pyramid organization of a WSI which consists of multiple images with different resolution at each level of the pyramid [1]. Initially, the slide is loaded into the memory at the top level of the pyramid which has a coarse resolution, and only a part of the WSI (a small tile) is read by the software. The right picture in the Fig. 2 illustrates that the WSI in each layer of the stack is consisted of many tiles. When doing the zoom-in operation, the bottom level of the WSI is loaded.
Whole-Slide imaging (WSI) had been the industry standard for the large-scale, high throughput digital pathology (DP). WSI can be shared to remote areas by the Internet and reviewed on the mobile devices. With the help of the Machine Learning and Deep Learning, DP can be applied in an automatic way.
5 steps for processing the WSI:
- Scale down the WSI into a small size.
- Filter the WSI and create a binary mask.
- Cut the WSI into tiles.
- Score the tiles (count the mitosis).
- create patches of the scored tiles.
Also, the WSI manipulation also needs specific libraies. To load and cut the region of the WSI, OpenSlide which is an open-source software is required. You can read this artical to know how to install and use OpenSlide.
After pre-processing, the WSI can be used for the further usage, for instance, fed into the Deep Learning model, statistics.
Reference:
[1] https://dicom.nema.org/dicom/dicomwsi/
[2] https://developer.ibm.com/articles/an-automatic-method-to-identify-tissues-from-big-whole-slide-images-pt1/